Introduction
Animals are regularly exposed to a wide diversity of bacteria with the potential to cause diseases. Many times, these bacteria can establish associations with other bacteria increasing the risk of infection. The interest in hemotrophic mycoplasmas (also called hemoplasmas) is growing worldwide, primarily because of their detection by molecular methods (Kaewmongkol et al., 2017; Maggi et al., 2013).
Hemoplasmas are Gram-negative pleomorphic bacteria, with small size (from 0.3 to 1 µm in diameter) and small genomes (0.5-1.0 Mb). They are considered obligate epierythrocytic bacteria, so far uncultivable and may act as opportunistic agents, that silently infect animals (Messick, 2004). Hemoplasmosis may cause hematological disorders in several mammalian species, ranging from severe anemia to chronic infection without clinical signs. Those animals with acute infections may present hemolysis, anorexia, dehydration, fever, loss of weight, lethargy and even sudden death (Messick, 2004;Willi et al., 2007). Fleas, ticks, lice, and flies are responsible for the transmission of hemoplasmas in cats, dogs, mice, pigs, and cattle (Hornok et al., 2008; Seneviratna et al., 19731973; Willi et al., 2010). Although hemoplasmosis is not strictly considered a tick-borne disease, ticks might play a role in the epidemiology of these bacteria in cattle since some hemoplasmas are occasionally detected in ticks (Barker et al., 2010).
In Mexico, the genome of bovine hemoplasmas Candidatus Mycoplasma haemobos and Mycoplasma wenyonii have been reported (Martínez-Ocampo et al., 2016; Quiroz-Castañeda et al., 2018) and the presence of Ca. M. haemobos was unknown until the first report of molecular detection of Ca. M. haemobos in 2018 (Jaimes-Martínez et al., 2018). However, the presence of Mycoplasma wenyonii had not been reported. In this work, we present the first molecular detection in blood bovine of M. wenyonii by molecular methods based of end-point PCR and the detection of Ca. M. haemobos and M. wenyonii in a duplex PCR reaction. In order to prevent dispersion of hemoplasmas in cattle, the development of diagnostic methods are necessary, especially when a limited number of hemoplasmas are suspected in animal blood.
1. Methodology
1.1. Genomic DNA extraction
A total of 27 blood samples were used to extract genomic DNA (gDNA). Four samples from Huitzilac, Morelos (9936, 9938, 9939, 9940), nine from Tlaquiltenango, Morelos (Prieta Ajuchitlán, 3313856, 1700366161, 1700158415, 1700366176, 1700158415, 1700366175, 1700158423, Cuerno mocho), nine form Tapalpa, Jalisco (4701, 1162, 6802, 2914, 13R, 6760, 6747, 4706, 2904) and five from the Germplasm bank of Anaplasma Unit (CENID-SAI, INIFAP) (MEX-15-099-01, MEX-31-096-01, MEX-17-017-01, 0705, 3668). Blood samples were stored at 4°C until genomic DNA extraction with the kit ReliaPrepTM Blood gDNA Miniprep System (Promega). The extracted gDNA was quantified by spectrophotometry (NABI) and then analyzed by electrophoresis in 1% agarose gel.
1.2. Detection of Candidatus Mycoplasma haemobos and Mycoplasma wenyonii by Polymerase Chain Reaction
To detect Ca. M. haemobos, a fragment of 492 bp of the 16S ribosomal RNA was amplified by PCR according to (A Girotto et al., 2012). The primer sequences are: Mhfwd: 5’-ATC TAA CAT GCC CCT CTG TA -3’ and Mhrev: 5’-GTA TTC GGT GCA AAC AA -3’. A fragment of 189 bp was amplified with the primers: Mwfwd: 5’-AGT CTG AGA TGA CTA TAG TG -3’ and Mwrev: 5’-CGA GGC AAA CCC CGC AAG CA -3’ that were designed based on the gene rnpB of M. wenyonii INIFAP02.
The PCR reaction contained 12.5 µL of MyTaq Mix 2X (Bioline), 1 µL of each primer (10 pmol/ µL), and 300 ng of gDNA as template and MiliQ water to a final volume of 25 µL. The PCR reaction conditions were: a first denaturation cycle at 94°C for three minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 2 minutes and extension at 72°C for 1 minute, finally one cycle at 72°C for 10 minutes. For detection of M. wenyonii (INIFAP02) the conditions were: a first denaturation cycle at 94°C for three minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 30 seconds and extension at 72°C for 30 seconds, finally one cycle at 72°C for 10 minutes. As negative control, DNA template was substituted by MilliQ water. After PCR amplification, samples were observed by electrophoresis in 1% agarose gel stained with ethidium bromide.
1.3. Duplex PCR for detection of Ca. M. haemobos and M. wenyonii
The gDNA of the positive samples to Ca. M. haemobos and M. wenyonii amplified by end-point PCR were used as templates for duplex reaction. This reaction amplifies both 492 bp and 189 bp fragments in the following conditions: 12.5 µL of MyTaq Mix 2X (Bioline), 1 µL of each primer (10 pmol/ µL), 300 ng of gDNA and MilliQ water to a final volume of 25 µL. Amplification conditions were: an initial denaturation at 94°C for three minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 2 minutes, extension at 72°C for 1 minuto and a last extension cycle at 72°C for 10 minutes. To test the sensitivity of the duplex PCR, 300 ng of gDNA from MEX-31-096-01 was used as template and diluted 1:10, 1:100, 1:1000 and 1:1000 (corresponding to 30 ng/µl, 3 ng/µl, 0.3 ng/µl and 0.03 ng/µl, respectively).
1.4. Purification of PCR products for sequencing
The amplicons of 492 bp and 189 bp were purified with the kit Wizard Gel and PCR Clean-Up System (Promega) and resuspended in 30 µL of MilliQ water and then sequenced in Unidad de Síntesis y Secuenciación, IBT-UNAM.
1.5. Propidium iodide staining
The blood infected with hemoplasmas were stained with propidium iodide. 7-10 µL of blood were smeared on a slide and then the blood was gently fixed drop by drop with cold methanol at room temperature. The sample was gently washed drop by drop three times with tridistilled water. Later, 15 µg (15 µL) of propidium iodide solution (stock 1 mg/ml) were mixed with 485 µL of PBS and then added drop by drop to the sample and dark-incubated for 10 minutes at room temperature. After incubation, the sample was washed three times with PBS as mentioned before and air-dried. The fluorescence of propidium iodide was observed in a epifluorescence microscope with an excitation/emission spectra of 493/636 nm.
2. Results
2.1. PCR amplification to detect Ca. M. haemobos and M. wenyonii
Both hemoplasmas were detected in the blood samples of bovines from different geographic sources. The amplicons obtained after end-point PCR are shown in Figure 1. Ca. M. haemobos was detected in 24 out of 25 blood samples (Figure 1A), while M. wenyonii was detected in 22 out of 27 blood samples (Figure 1B). The sequencing of amplicons and the multiple alignment of sequences (Clustal Omega) confirmed that the amplicon of 189 bp have a 100% of identity with M. wenyonii INIFAP02 while the amplicon of 492 bp have a range of identity of 96-100% Ca. M. haemobos INIFAP01.
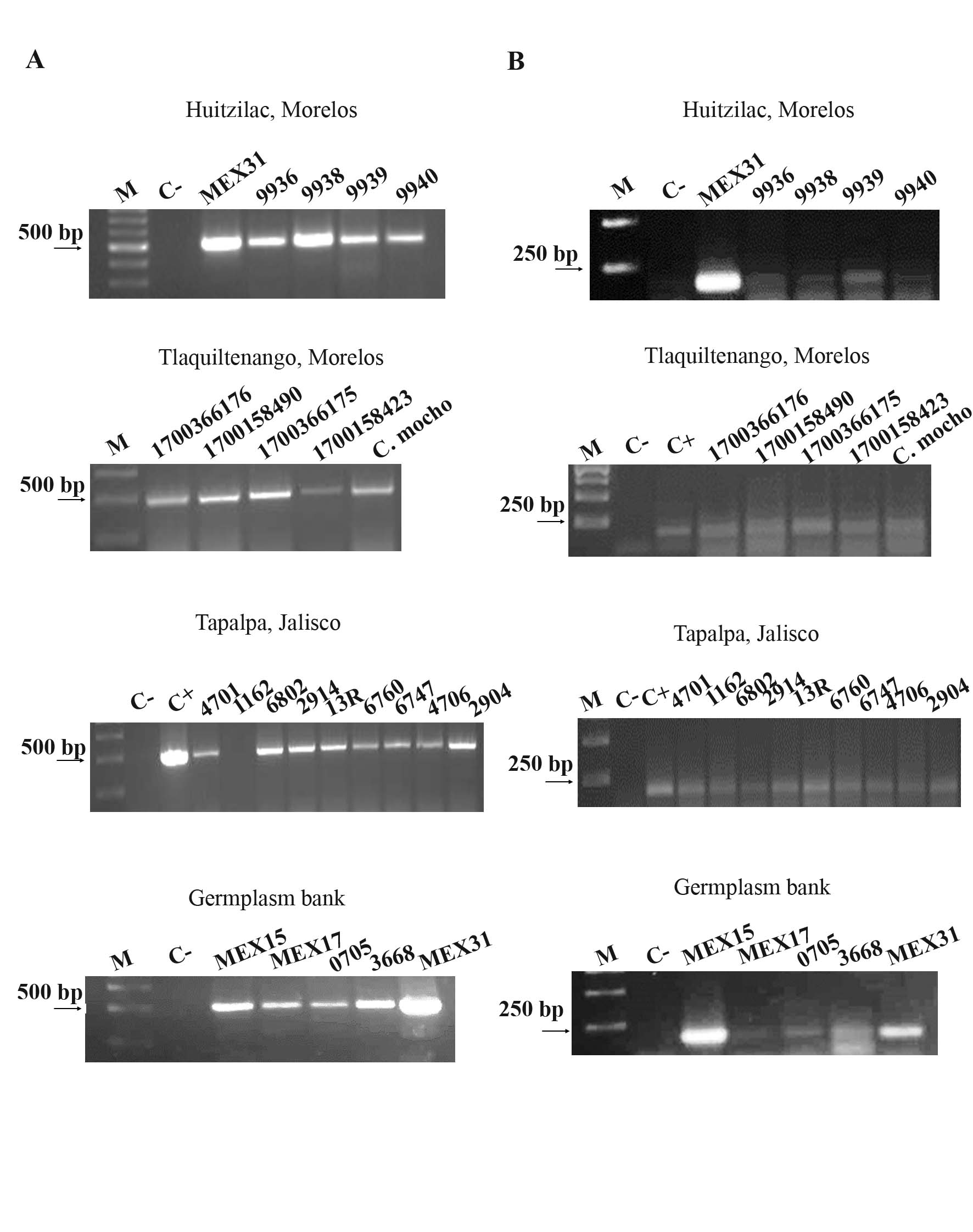 |
Molecular detection of Ca. M. haemobos and M. wenyonii. A) PCR amplification of the fragment of 492 bp of 16S rRNA of Ca. M. haemobos. This hemoplasma was detected in 24 out of 25 blood samples. B) PCR amplification of the fragment of 189 bp of rnpB gene of M. wenyonii detected in 26 out of 27 blood samples. M, molecular marker, C-, negative reaction control. |
2.2. Duplex PCR amplification to detect Ca. M. haemobos and M. wenyonii
Some gDNA samples that were positive to Ca. M. haemobos and M. wenyonii (MEX31, 1162, 4701, 6802, 2914, 2904, 1700366176, 1700158490, 1700366175) were used to perform the duplex PCR. Both ampicons of 189 bp and 492 bp were detected in 5 out of 10 gDNA samples as is shown in Figure 2.
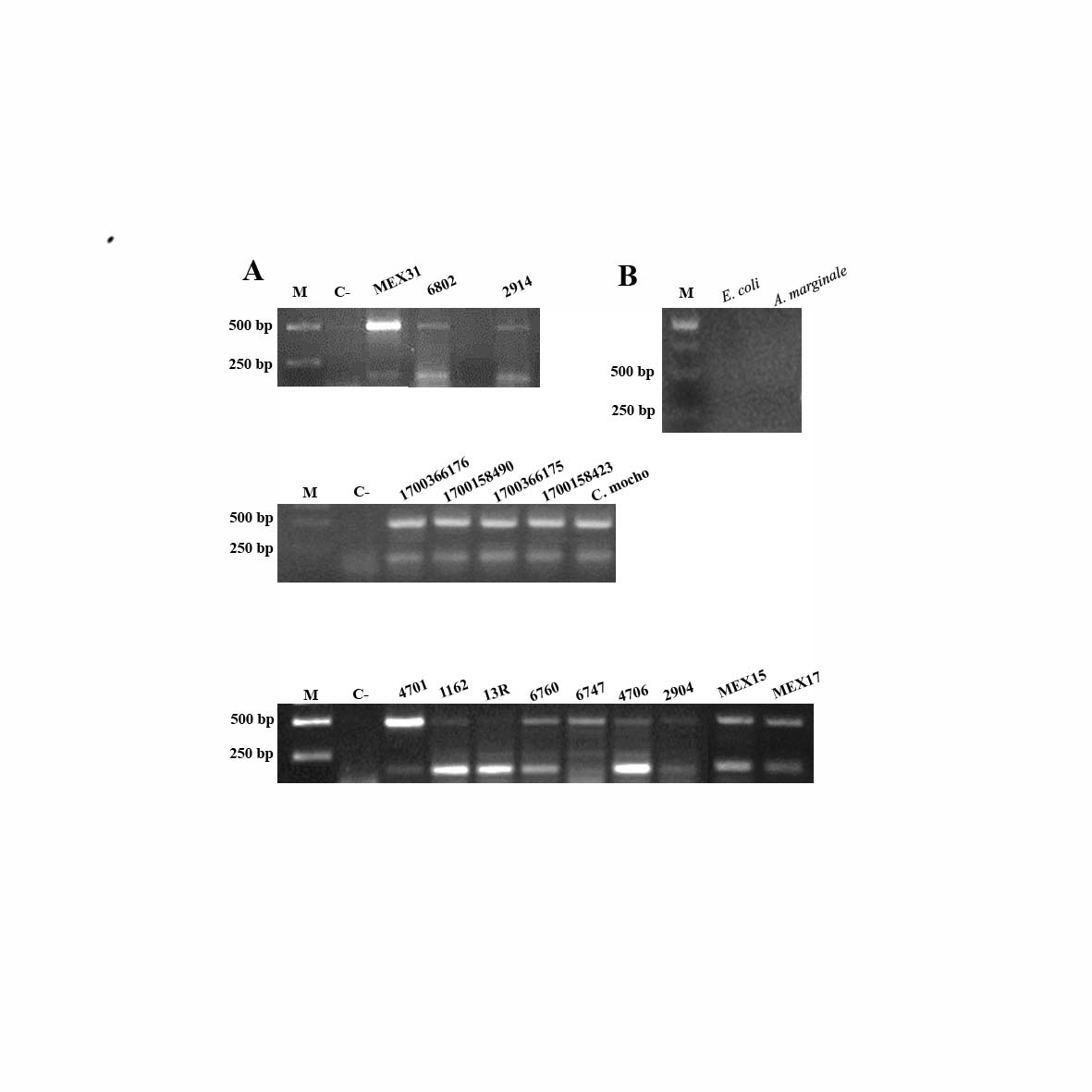 |
Duplex PCR to detect hemoplasmas. A) Amplicons of 189 bp and 492 bp detected by duplex PCR in 17 blood samples (MEX31, 6802, 2914, 1700366176, 1700158490, 1700366175, 1700158423, C. mocho, 4701,1162, 13R, 6760, 6747, 4706, 2904, MEX15, MEX17. B) Specificity of the duplex PCR, the genomic DNA used as template was from Escherichia coli and A. marginale, both, considered as pathogens of cattle. M, molecular marker, C-, negative reaction control. |
2.3. Sensitivity of duplex PCR
The minumun detectable amount of gDNA of MEX-31-096-01 used in duplex PCR was in the dilution 1:10000, this is, 0.03 ng/µl. Figure 3 shows that both amplicons are detected in dilutions 1:10, 1:100 and 1:1000. In dilution 1:10000 only the amplicon of 492 bp was detected.
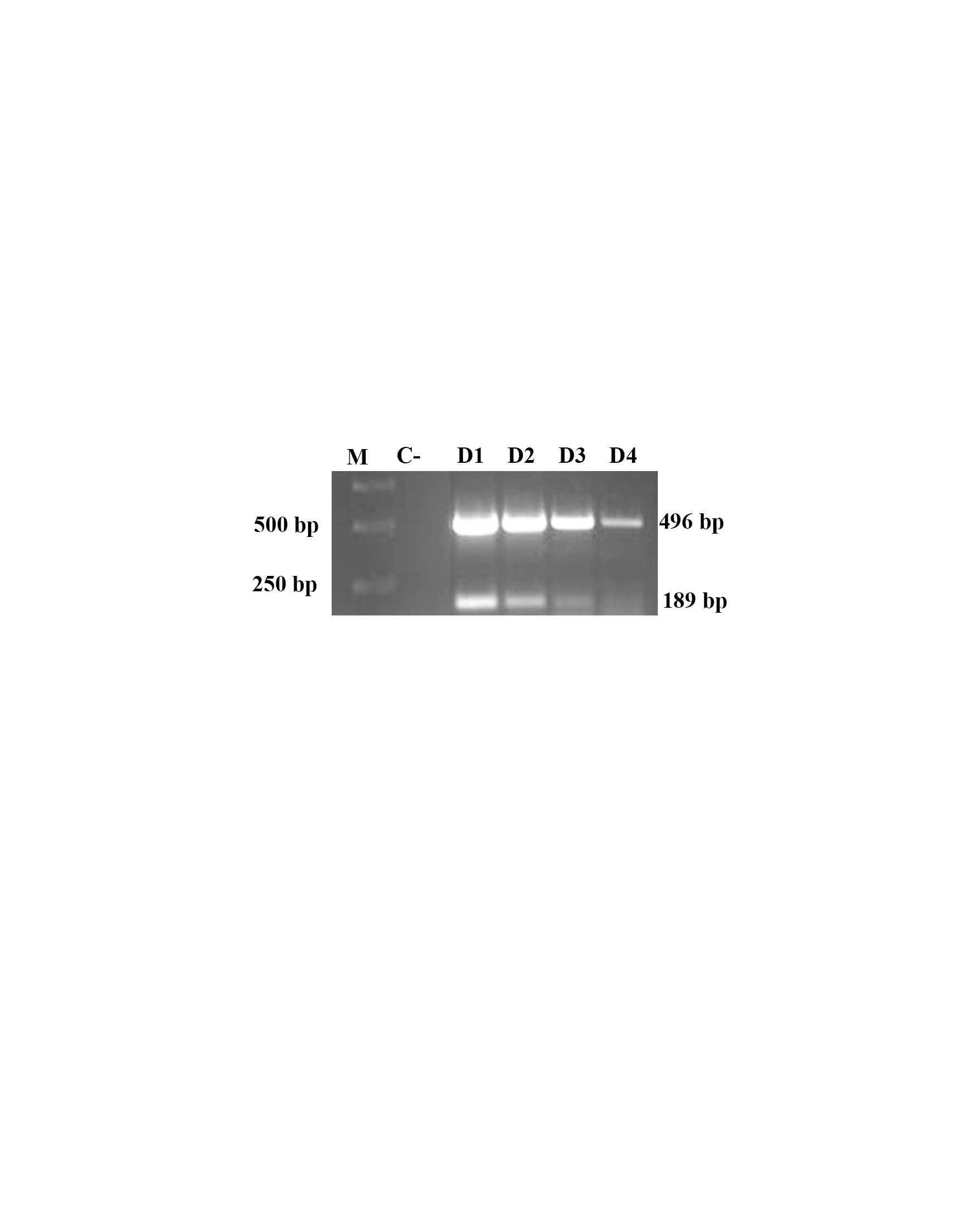 |
Sensitivity of duplex PCR. Both amplicons (189 bp and 492 bp) were detected in dilutions D1, D2 and D3 of gDNA from blood infected with Ca. M. haemobos and M. wenyonii. (D1, Dilution 1:10, D2, Dilution 1:100; D3, dilution 1:1000; and D4, dilution 1:10000), M, molecular marker, C-, negative reaction control. |
2.4. Propidium iodide staining of hemoplasmas
Additionally to molecular detection of hemoplasmas, they were stained with propidium iodide to confirm microscopically their presence as epierythrocytic bacteria. We found that hemoplasmas are located outside the erythrocyte as has been reported as hemoplasmas, as is shown in Figure 4.
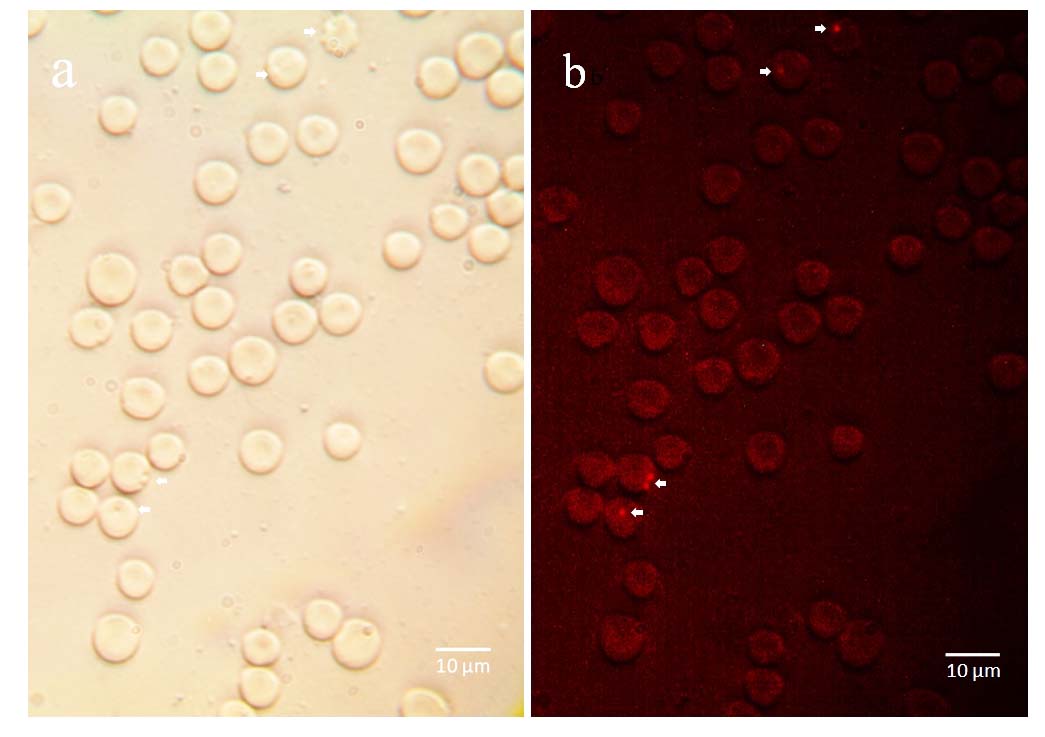 |
Representative micrograph of hemoplasmas stained with propidium iodide. M. wenyonii was detected as epierythrocytic pathogen. a) bright field of erythrocytes infected with hemoplasmas; b) hemoplasmas stained with propidium iodide are observed outside the erythrocytes. |
3. Discusion
Bovine hemoplasmas, Ca. M. haemobos and M. wenyonii have been detected in several countries around the world. In Latin America, they were reported in Mexico, Brazil and Cuba (Díaz-Sánchez et al., 2019; A Girotto et al., 2012; Jaimes-Martínez et al., 2018). The risk of coinfections of these hemoplasmas with other pathogens endanger the health of cattle and causes economical losses. In 2018, Ca. M. haemobos was detected in Mexican cattle by molecular methods, and sometimes this hemoplasma was found in co-infection with pathogen A. marginale (Jaimes-Martínez et al., 2018). Due to hemoplasmas can cause anemia, loss of weight, fever, edema, reproductive problems, mastitis and jaundice, their detection is the first step to characterize the agents present in the infected host (Ayling et al., 2012). Today, the use of molecular markers and their PCR amplification is the most accessible and common method to detect the presence of pathogens. Here, we report the molecular detection of hemoplasmas in blood samples from Mexican cattle. The blood samples come from different geographical locations, including, Morelos (Huitzilac, Tlaquiltenango, MEX17), Jalisco (Tapalpa), Estado de México (MEX15) and Yucatán (MEX31). The presence of these hemoplasmas in these samples suggest that they are spread in our country. Nevertheless, a study with more samples is still required, especially in those states with a significant impact in cattle production as Veracruz, Tamaulipas, Sonora, Guanajuato and Michoacán. The detection of each hemoplasma (either Ca. M. haemobos or M. wenyonii) was based on end-point PCR while a duplex PCR detected both hemoplasmas. We found that amplification by PCR of the molecular marker 16S ribosomal RNA gene allowed the detection of Ca. M. haemobos in 96% of the blood samples analyzed. The rnpB gene was successfully used to detect M. wenyonii in 95.29% of the blood samples, as it was confirmed by sequencing identity (figure 1).
Both hemoplasmas were detected by duplex PCR in 17 blood samples (figure 2A), the design of the primers was specific to hemoplasmas, since amplicons were not observed in samples with gDNA from E. coli and A. marginale (figure 2B). Altough the development of detection methods based on duplex PCR requires optimization of primer annealing conditions, and enzyme and buffer concentrations for maximum amplification efficiency of each gene target, in this work, we were able to design a PCR reaction with two pairs of primers to detect hemoplasmas in cattle, in samples with as little as 0.03 ng of gDNA (figure 3). The hemoplasmas stained with propidium iodide were observed on the surface of the erythrocytes (figure 4).
This is the first report of the detection of hemoplasmas in Mexican cattle based on end-point PCR and the detection of Ca. M. haemobos and M. wenyonii in a duplex PCR. The conditions of duplex PCR allowed the detection of both hemoplasmas in 17 samples. With this approach to detect bovine hemoplasmas in Mexican cattle, veterinarians and authorities may could be able to take decisions to avoid the spread of pathogens in cattle and decrese the risk of coinfections that may affect animal health.
4. Conclusions
Hemoplasmas found either alone or in co-infections become a risk to animal health. Hence, the importance to develop precise molecular tools for early detection of pathogens in cattle. In this work, we show the detection of both hemoplasmas that affect cattle by molecular methods.
Acknowledgements
To Ms. C. Elizabeth Salinas Estrella for kindly provide some blood samples.